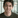Running DVC on a SLURM cluster
For many ML projects, there comes a point when local development hits the wall and we need to scale up the underlying compute resources. Maybe the dataset grows too large for your primary workstation or the deep learning model requires several high-end GPUs. This should be a routine transition for ML developers, and one to which they shouldn’t have to give much thought. In this blog post, we’ll explain our approach to remote DVC experiments on a SLURM cluster and share some code to get you started with the same.

Introduction
For many ML projects, there comes a point when local development hits the wall and we need to scale up the underlying compute resources. Maybe the dataset grows too large for your primary workstation or the deep learning model requires several high-end GPUs. This should be a routine transition for ML developers, and one to which they shouldn’t have to give much thought. In this blog post, we’ll explain our approach to remote DVC experiments on a SLURM cluster and share some code to get you started.
We work at an AI-driven precision medicine company called Exscientia. Our goal is to change the way the world discovers and develops new medicines. The company is roughly evenly split between biologists and chemists on one side and technologists on the other, with your two authors belonging to the latter group; Dom is an AI research scientist and Luis is an engineer. This context is important to understand why we gravitated towards DVC in the first place, and why we scaled it up the way we did.
Why DVC on SLURM?
As demonstrated in research undertaken by the DevOps movement, it’s hard to maintain consistent software delivery without well-designed tooling (like CI/CD) and a conducive developer culture (like PRs or working in small batches). Our domain is highly specific, but the same principles apply: to move fast while maintaining high quality, reliability and reproducibility, we need to adopt best DevOps practices. There are only so many hours in a day and you want to spend all of them on trying out new ideas and ideally none on setting up infrastructure. Good tooling optimises scientists’ efficiency and lets them run more experiments, each more thorough and exhaustive than would otherwise have been possible – all this while maintaining control over research code bases which can, if left unchecked, turn into precarious Jenga towers. Predictable code with clear standards also eases collaboration, the lifeblood of science. Consequently it’s much more important to pick an arbitrary standard than to obsess over any particular detail.
At Exscientia we provide researchers with project templates that automatically set up version control and CI/CD as well as QA tooling like Black, Ruff and Mypy. To coherently extend this setup to the joint realms of data science and ML, we integrated DVC. Our scientists can set up a fresh DVC-enabled repository with all the productivity tooling in just a few keystrokes and start experimenting right away. And because DVC transparently extends Git, there is less tool-induced context switching: users are always dealing with Git in some shape or form, rather than Git (for the code) and a database hidden behind a web service (for all the rest of it). Less context switching translates to less frustration and more flow.
To maintain a frictionless developer experience even as model sizes grow beyond the means of the humble laptop, we surveyed the organisation’s entire computational estate with a view towards designing an effective developer experience. Our platforms must support a number of teams with on-demand Jupyter or RStudio instances as well as workflow orchestration engines. We need to run large unsupervised jobs, interactive analyses and development sessions across many domains and technologies: data processing, ML model training and chemical simulations, each with different resource requirements. Finally, submitting a large workload should be a smooth and routine experience.
In the end, a cloud-deployed SLURM cluster fit the bill. It can efficiently scale compute resources while maintaining a user-friendly interface for job submission. As a bonus, many of our users are already familiar with SLURM from their past lives in academia. The principal mode of interaction is very simple: the user submits a Bash script describing exactly what they want to happen, including the exact resources required. SLURM will wait until such resources are available and then execute the job as instructed. Thanks to this highly general interface, the same computational resource, and its administrators, can support very diverse groups of users at the same time, reducing infrastructural complexity across the organisation.
A sample project
We’ll set up a basic project for this demo and, to keep with the drug discovery theme, we will be predicting solubility of chemical compounds in water using only our recently open-sourced framework MolFlux.
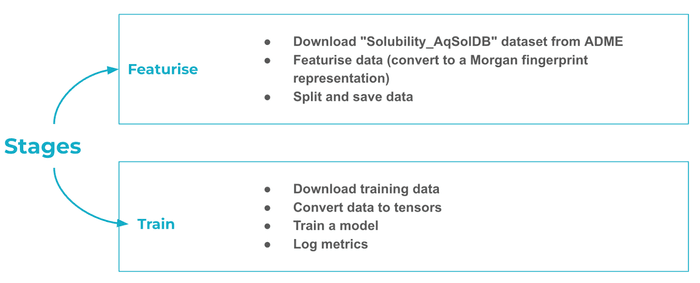
The DVC pipeline consists of a featurisation stage, which loads the “ESOL” dataset consisting of pairs of molecules and their aqueous solubilities - how easily a molecule dissolves in water.
A few words about molecules and neural networks. Cheminformatics typically represents molecules as graphs, with atoms acting as the nodes and chemical bonds as the edges. There are several ways to feed molecular data to neural networks, each with its own pros and cons. GNNs can act directly on the molecular graph. You can also represent the graph as a string (most commonly using the SMILES format) and feed it to any sequence model such as a transformer.
In this example we’ll use a classic cheminformatics transformation called ECFP, or extended connectivity fingerprint. It’s essentially analogous to n-grams in NLP, which track whether a particular sequence of tokens appears in a text document. For example, does the 3-letter sequence “wea” appear in the Wikipedia article on blazers? Indeed it does, as part of “wear”.
Returning to ECFPs defined on molecular graphs, each “n-gram” is an atom and its immediate (e.g. 2-hop) neighbourhood. Since the “vocabulary” of all possible “n-grams” is finite, we can associate to each molecule a finite bit-vector (of the same length as the vocabulary) such that the choice of 0 or 1 indicates whether the corresponding “n-gram” is present in the molecule. This bit-vector is the ECFP fingerprint. And since it has a constant length, we can feed it into a large variety of ML algorithms, such as the MLP in the training stage.
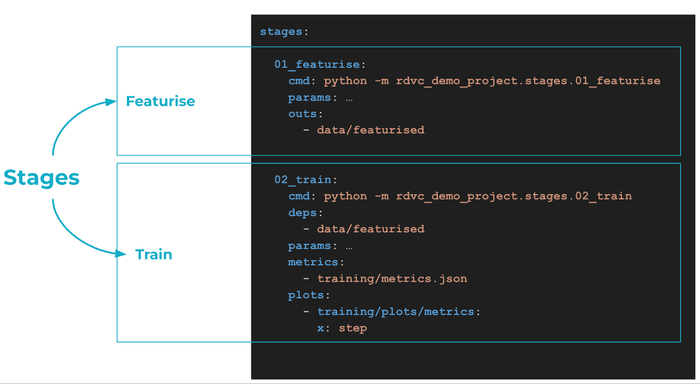
We use DVC to configure and run the pipeline, decoupling the data featurisation step (where we convert molecules to ECFPs) from the model training step.
DVC pipelines are useful to organise projects. As they are versioned in Git, you can reproduce complete workflows and results. Running a new experiment is a command away:
$ dvc exp runThis executes and tracks experiments in your repository without polluting it with unnecessary Git commits, branches, directories, etc. For more information and examples, see the DVC documentation.
It may not be immediately obvious, but our setup is highly modular. Head over to
src/rdvc_demo_project/config/main.yaml to see just an example of configuration
options we can tweak for each individual experiment. To start a much longer
training run, execute
$ dvc exp run -S model.config.trainer.max_epochs=100MolFlux was built to be explicitly config-driven and DVC’s Hydra integration exposes all of that flexibility out of the box.
In the cloud
Now that DVC experiments run on our local machine, we’d like to move them to the
SLURM cluster. In this second repository, we share the source code to an
internal tool we call rDVC (for remote
DVC). It is, by design, a very thin layer around dvc exp run and accepts all
of its options and arguments. But on top of that it also recognises many of
sbatch arguments and flags, allowing
it to control which computational resource inside the cluster will be used and
for how long. For a full list of options consult rdvc run –help.
Let’s demonstrate how it works.
On its own, DVC knows nothing about your remote cluster, so we’ll need to start
with a small amount of setup. Make sure you have cloned the sample project repo
and installed the Python virtual environment using init_python_venv.sh. You
will initialise your local rDVC config with
$ rdvc init projectFollow the wizard to set up default options for this project’s remote runs; they
will be found in .rdvc/config.toml inside of the project repository. Depending
on the cluster’s setup, you may be able to choose the instance type allocated
to your job. For the demo we have configured the cluster with t3.xlarge,
g5.xlarge and g5.12xlarge. Our internal version of rDVC supports many more
instance types and we encourage you to fork rDVC, redefine supported instance
types and make the tool your own. For this demo, we pick g5.xlarge as the
default instance as we want access to the GPU. But let’s continue with the demo.
To point rDVC at your SLURM cluster, we’ll run the global initialisation script
next:
$ rdvc init globalrDVC now knows how to contact SLURM, so let’s finish with configuration of the remote server:
$ rdvc init remoteNothing stands between us and a remote GPU-powered experiment! Since rDVC is in
many ways just a wrapper around dvc exp run, we can easily set off a run with
modified parameters as
$ rdvc run -S fabric=gpuWhen your run is finished you can pull it to your local repository with
$ dvc exp pull originand look at the results.
Behind the scenes
rDVC compiled a SLURM batch (or “sbatch”) script containing these instructions:
- Clone the project repo
#!/bin/bash
#SBATCH --output=".rdvc/logs/slurm-%j.out"
#SBATCH --job-name=rdvc-run:rdvc-demo-project:main
#SBATCH --wckey=rdvc-demo-project
#SBATCH --mail-type=END,FAIL
#SBATCH --mail-user=<[email protected]>
#SBATCH --constraint=t3.xlarge
#SBATCH --cpus-per-task=2
#SBATCH --nodes=1
#SBATCH --exclusive
# Ensure bashrc is loaded
source "${HOME}/.bashrc"
# Exit on failure http://redsymbol.net/articles/unofficial-bash-strict-mode/
set -euxo pipefail
IFS=$'\n\t'
export RDVC_JOB_REPO_NAME="rdvc-demo-project"
export RDVC_JOB_REPO_URL="[email protected]:<user>/rdvc-demo-project.git"
export RDVC_JOB_REPO_BRANCH="main"
export RDVC_JOB_REPO_REV="<git_hash>"
export RDVC_DIR="${RDVC_DIR:-${HOME}/.rdvc}"
# Prepare a directory for the current job
export RDVC_JOB_WORKSPACE_DIR="/tmp/rdvc-${SLURM_JOB_ID}"
mkdir -p "${RDVC_JOB_WORKSPACE_DIR}"
# Ensure cleanup after job finishes, regardless of exit status
function cleanup_job_dir(){
echo "Cleaning up the job directory."
rm -rf "${RDVC_JOB_WORKSPACE_DIR}"
}
trap cleanup_job_dir EXIT
# Create an insulated Git workspace for the current job
echo "Creating Git workspace."
export RDVC_JOB_REPO_DIR="${RDVC_JOB_WORKSPACE_DIR}/${RDVC_JOB_REPO_NAME}"
git clone --branch "${RDVC_JOB_REPO_BRANCH}" "${RDVC_JOB_REPO_URL}" "${RDVC_JOB_REPO_DIR}"
cd "${RDVC_JOB_REPO_DIR}" || exit
# Ensure the job runs on the same revision as was submitted (even if the branch has moved on in the meantime)
git checkout "${RDVC_JOB_REPO_REV}"- Install the Python virtual environment with
init_python_venv.sh
# Install Python environment
echo "Install Python environment."
./init_python_venv.sh
echo "Activate Python environment."
source ./.venv/bin/activate
# Setup links for the DVC cache shared among jobs and projects
dvc config --local cache.type hardlink,symlink,copy
# Push results of experiments even if job fails
function cleanup_dvc(){
if [ "$1" != "0" ]; then
# Push cache of all runs, including failed
echo "Job failed. Pushing run cache."
dvc push --run-cache
else
echo "Job successfully finished."
fi
deactivate
cleanup_job_dir
}
trap 'cleanup_dvc $?' EXIT- Execute dvc exp run -S fabric=gpu
export RDVC_JOB_EXP_RUN_OPTIONS_STRING="-S fabric=gpu"
echo "Executing DVC experiment."
eval "dvc exp run --pull --allow-missing ${RDVC_JOB_EXP_RUN_OPTIONS_STRING}"- Push the experiment to the remote
# Push experiment to the remote and update the repository
echo "Pushing DVC experiment to Git and DVC remotes."
dvc exp push $RDVC_JOB_REPO_URLThis script is submitted to the cluster over SSH. You can view it in
~/.rdvc/submissions.
And that’s it! It’s so simple you could do it manually in an interactive SLURM
session - and that happens to be a good way to debug issues. If your job fails,
first consult its log over at ~/.rdvc/logs and then try to reproduce the
submission script from an interactive session.
Conclusion
We shared two repositories: a simple DVC project and a tool for remote execution on SLURM clusters. The latter is universal - it knows nothing about the project! - and easily hackable. We highly recommend to fork and customise it to your team’s needs.